Standard Plate Count Microbiology Results Beef
BAM Chapter 3: Aerobic Plate Count
Jan 2001
Authors: Larry Maturin (ret.) and James T. Peeler (ret)
For additional information, contact Guodong Zhang.
The aerobic plate count (APC) is intended to indicate the level of microorganism in a product. Detailed procedures for determining the APC of foods accept been developed by the Clan of Official Belittling Chemists (AOAC) (3) and the American Public Wellness Association (APHA) (1). The conventional plate count method for examining frozen, chilled, precooked, or prepared foods, outlined below, conforms to AOAC Official Methods of Assay, sec. 966.23, with one procedural change (966.23C). The suitable colony counting range (x) is 25-250. The automated spiral plate count method for the examination of foods and cosmetics (v), outlined below, conforms to AOAC Official Methods of Analysis, sec. 977.27. For procedural details of the standard plate count, see ref. ii.Guidelines for calculating and reporting plate counts take been inverse to conform with the anticipated changes in the 16th edition of Standard Methods for the Examination of Dairy Products (two) and the International Dairy Federation (IDF) procedures (six).
- Conventional Plate Count Method
- Spiral Plate Method
- References
Conventional Plate Count Method
- Equipment and materials
- Work area, level tabular array with ample surface in room that is clean, well-lighted (100 foot-candles at working surface) and well-ventilated, and reasonably gratuitous of dust and drafts. The microbial density of air in working area, measured in fallout pour plates taken during plating, should not exceed 15 colonies/plate during 15 min exposure.
- Storage infinite, gratis of grit and insects and adequate for protection of equipment and supplies
- Petri dishes, glass or plastic (at to the lowest degree 15 × xc mm)
- Pipets with pipet aids (no oral cavity pipetting) or pipettors, i, 5, and ten ml, graduated in 0.one ml units
- Dilution bottles, 6 oz (160 ml), borosilicate-resistant glass, with safe stoppers or plastic screw caps
- Pipet and petri dish containers, adequate for protection
- Circulating h2o bath, for tempering agar, thermostatically controlled to 45 ± 1°C
- Incubator, 35 ± 1°C; milk, 32 ± one°C
- Colony counter, dark-field, Quebec, or equivalent, with suitable light source and grid plate
- Tally register
- Dilution blanks, 90 ± i ml Butterfield's phosphate-buffered dilution water (R11); milk, 99 ± 2 ml
- Plate count agar (standard methods) (M124)
- Refrigerator, to absurd and maintain samples at 0-5°C; milk, 0-4.4°C
- Freezer, to maintain frozen samples from -15 to -xx°C
- Thermometers (mercury) appropriate range; accuracy checked with a thermometer certified past the National Institute of Standards and Technology (NIST)
- Procedure for analysis of frozen, chilled, precooked, or prepared foods
Using separate sterile pipets, ready decimal dilutions of 10-2, 10-iii, 10-four, and others equally appropriate, of food homogenate (see Chapter i for sample grooming) by transferring 10 ml of previous dilution to 90 ml of diluent. Avert sampling cream. Shake all dilutions 25 times in 30 cm (1 ft) arc within vii s. Pipet 1 ml of each dilution into split up, duplicate, appropriately marked petri dishes. Reshake dilution bottle 25 times in 30 cm arc within 7 s if it stands more than iii min before information technology is pipetted into petri dish. Add 12-15 ml plate count agar (cooled to 45 ± i°C) to each plate within 15 min of original dilution. For milk samples, pour an agar control, cascade a dilution water command and pipet h2o for a pipet control. Add together agar to the latter two for each series of samples. Add together agar immediately to petri dishes when sample diluent contains hygroscopic materials, due east.thou., flour and starch. Pour agar and dilution water control plates for each serial of samples. Immediately mix sample dilutions and agar medium thoroughly and uniformly past alternate rotation and back-and-forth motility of plates on apartment level surface. Allow agar solidify. Invert solidified petri dishes, and incubate promptly for 48 ± 2 h at 35°C. Practice not stack plates when pouring agar or when agar is solidifying.
- Guidelines for computing and reporting APCs in uncommon cases
Official Methods of Analysis (3) does not provide guidelines for counting and reporting plate counts, whereas Standard Methods for the Examination of Dairy Products, 16th ed. (2) presents detailed guidelines; for uniformity, therefore, apply APHA guidelines every bit modified (6,8). Written report all aerobic plate counts (2) computed from duplicate plates. For milk samples, report all aerobic plate (2) counts computed from indistinguishable plates containing less than 25 colonies as less than 25 estimated count. Report all aerobic plate counts (2) computed from duplicate plates containing more than than 250 colonies as estimated counts. Counts outside the normal 25-250 range may give erroneous indications of the actual bacterial composition of the sample. Dilution factors may exaggerate low counts (less than 25), and crowded plates (greater than 250) may be difficult to count or may inhibit the growth of some leaner, resulting in a low count. Report counts less than 25 or more than 250 colonies as estimated aerobic plate counts (EAPC). Use the following guide:
- Normal plates (25-250). Select spreader-free plate(s). Count all colony forming units (CFU), including those of pinpoint size, on selected plate(due south). Record dilution(due south) used and full number of colonies counted.
- Plates with more than than 250 colonies. When number of CFU per plate exceeds 250, for all dilutions, tape the counts as likewise numerous to count (TNTC) for all but the plate closest to 250, and count CFU in those portions of plate that are representative of colony distribution. See ref. 2 for detailed guidelines. Marking calculated APC with EAPC to denote that it was estimated from counts exterior 25-250 per plate range (see D-3).
- Spreaders. Spreading colonies are usually of three distinct types: ane) a chain of colonies, non too distinctly separated, that appears to be caused past disintegration of a bacterial clump; 2) one that develops in film of water between agar and bottom of dish; and iii) ane that forms in moving-picture show of h2o at edge or on surface of agar. If plates prepared from sample have excessive spreader growth and then that (a) area covered past spreaders, including total area of repressed growth, exceeds 50% of plate area, or (b) expanse of repressed growth exceeds 25% of plate expanse, report plates as spreaders. When it is necessary to count plates containing spreaders non eliminated past (a) or (b) above, count each of the 3 distinct spreader types as one source. For the beginning blazon, if merely one chain exists, count it every bit a single colony. If i or more than bondage appear to originate from divide sources, count each source as 1 colony. Practice not count each individual growth in such bondage equally a separate colony. Types 2 and 3 usually result in distinct colonies and are counted equally such. Combine the spreader count and the colony count to compute the APC.
- Plates with no CFU. When plates from all dilutions take no colonies, report APC as less than 1 times the corresponding lowest dilution used. Mark calculated APC with asterisk to denote that it was estimated from counts outside the 25-250 per plate range. When plate(s) from a sample are known to be contaminated or otherwise unsatisfactory, record the effect(s) as laboratory blow (LA).
-
Calculating and recording counts (see refs vi, 8)
To avoid creating a fictitious impression of precision and accuracy when computing APC, report only the first 2 significant digits. Circular off to two meaning figures only at the fourth dimension of conversion to SPC. For milk samples, when plates for all dilutions accept no colonies, report APC equally less than 25 colonies estimated count. Round past raising the second digit to the next highest number when the third digit is 6, 7, eight, or nine and use zeros for each successive digit toward the right from the 2d digit. Round down when the third digit is 1, 2, 3, or 4. When the third digit is 5, round upwardly when the 2d digit is odd and round down when the second digit is even.
Examples
Calculated Count APC 12,700 13,000 12,400 12,000 15,500 xvi,000 fourteen,500 xiv,000 - Plates with 25-250 CFU.
- Calculate the APC as follows:
where N = Number of colonies per ml or g of production
Σ c = Sum of all colonies on all plates counted
due north1 = Number of plates in first dilution counted
north2 = Number of plates in second dilution counted
d = Dilution from which the first counts were obtainedInstance
1:100 1:1000 232, 244 33, 28 = 537/0.022
= 24,409
≈ 24,000 - When counts of duplicate plates fall within and without the 25-250 colony range, use just those counts that fall inside this range.
- Calculate the APC as follows:
-
All plates with fewer than 25 CFU. When plates from both dilutions yield fewer than 25 CFU each, record actual plate count but tape the count as less than 25 × ane/d when d is the dilution gene for the dilution from which the first counts were obtained.
Case
Colonies 1:100 one:chiliad EAPC/ml (g) eighteen 2 <ii,500 0 0 <ii,500 -
All plates with more than 250 CFU. When plates from both 2 dilutions yield more than 250 CFU each (just fewer than 100/cmtwo), guess the aerobic counts from the plates (EAPC) nearest 250 and multiply by the dilution.
Case
Colonies 1:100 1:m EAPC/ml (g) TNTC 640 640,000 TNTC, too numerous to count.
EAPC, estimated aerobic plate count. -
All plates with spreaders and/or laboratory accident. Report respectively as Spreader (SPR), or Laboratory Accident (LA).
-
All plates with more than than an average of 100 CFU per sq cm. Guess the APC equally greater than 100 times the highest dilution plated, times the area of the plate. The examples below accept an average count of 110 per sq cm.
Instance
Colonies/Dilution 1:100 ane:1000 EAPC/ml (g) TNTC 7,150(a) >6,500,000 EAPC(b) TNTC 6,490 >v,900,000 EAPC a Based on plate area of 65 cmii
b EAPC, estimated APC
c Based on plate area of 59 cm2
- Plates with 25-250 CFU.
Screw Plate Method
The spiral plate count (SPLC) method for microorganisms in milk, foods, and cosmetics is an official method of the APHA (2) and the AOAC (iii). In this method, a mechanical plater inoculates a rotating agar plate with liquid sample. The sample book dispensed decreases equally the dispensing stylus moves from the center to the edge of the rotating plate. The microbial concentration is adamant by counting the colonies on a part of the petri dish where they are easily countable and dividing this count by the appropriate volume. 1 inoculation determines microbial densities between 500 and 500,000 microorganisms/ml. Additional dilutions may be made for suspected high microbial concentrations.
- Equipment and materials
- Screw plater (Spiral Systems Instruments, Inc., 7830 Old Georgetown Road, Bethesda, Dr. 20814)
- Spiral colony counter (Screw Systems) with special grid for relating deposited sample volumes to specific portions of petri dishes
- Vacuum trap for disposal of liquids (2-4 liter vacuum bottle to deed every bit vacuum reservoir and vacuum source of 50-threescore cm Hg)
- Disposable micro beakers, 5 ml
- Petri dishes, plastic or glass, 150 × 15 mm or 100 × fifteen mm
- Plate count agar (standard methods) (M124)
- Calculator (optional), inexpensive electronic hand computer is recommended
- Polyethylene bags for storing prepared plates
- Commercial sodium hypochlorite solution, about 5% NaOCl (bleach)
- Sterile dilution water
- Syringe, with Luer tip for obstructions in stylus; chapters not critical
- Piece of work area, storage infinite, refrigerator, thermometers, tally, incubator, as described for Conventional Plate Count Method, above.
- Sodium hypochlorite solution (five.25%). Available commercially.
- Preparation of agar plates.
Automated dispenser with sterile delivery system is recommended to set agar plates. Agar volume dispensed into plates is reproducible and contamination charge per unit is low compared to hand-pouring of agar in open laboratory. When possible, use laminar air menstruum hood along with automated dispenser. Pour aforementioned quantity of agar into all plates so that aforementioned height of agar will exist presented to spiral plater stylus tip to maintain contact bending. Agar plates should be level during cooling.
The following method is suggested for prepouring agar plates: Apply automatic dispenser or pour constant amount (about fifteen ml/100 mm plate; l ml/150 mm plate) of sterile agar at threescore-70°C into each petri dish. Let agar solidify on level surface with poured plates stacked no higher than x dishes. Place solidified agar plates in polyethylene numberless, close with ties or heat-sealer, and shop inverted at 0-4.four°C. Bring prepoured plates to room temperature before inoculation.
- Training of samples.
As described in Chapter ane, select that role of sample with smallest amount of connective tissues or fatty globules.
- Description of spiral plater.
Spiral plater inoculates surface of prepared agar plate to permit enumeration of microorganisms in solutions containing between 500 and 500,000 microorganisms per ml. Operator with minimum training can inoculate 50 plates per h. Within range stated, dilution bottles or pipets and other auxiliary equipment are not required. Required bench space is minimal, and fourth dimension to check instrument alignment is less than 2 min. Plater deposits decreasing corporeality of sample in Archimedean spiral on surface of prepoured agar plate. Volume of sample on any portion of plate is known. After incubation, colonies appear along line of spiral. If colonies on a portion of plate are sufficiently spaced from each other, count them on special grid which associates a calibrated volume with each expanse. Guess number of microorganisms in sample past dividing number of colonies in a defined area by volume independent in same area. Studies have shown the method to be skillful not merely with milk (4) but as well with other foods (vii,10).
- Plating procedure
Check stylus tip angle daily and accommodate if necessary. (Utilise vacuum to concord microscope cover slip against face of stylus tip; if cover slip aeroplane is parallel at about l mm from surface of platform, tip is properly oriented). Liquids are moved through system past vacuum. Clean stylus tip by rinsing for 1 south with sodium hypochlorite solution followed by sterile dilution water for 1 s earlier sample introduction. This rinse procedure betwixt processing of each sample minimizes cantankerous-contamination. After rinsing, describe sample into tip of Teflon tubing by vacuum practical to 2-way valve. When tubing and syringe are filled with sample, close valve fastened to syringe. Identify agar plate on platform, identify stylus tip on agar surface, and kickoff motor. During inoculation, label petri plate lid. After agar has been inoculated, stylus lifts from agar surface and spiral plater automatically stops. Remove inoculated plate from platform and cover information technology. Move stylus back to starting position. Vacuum-rinse system with hypochlorite and water, and then innovate new sample. Invert plates and promptly place them in incubator for 48 ± 3 h at 35 ± 1°C.
- Sterility controls
Check sterility of spiral plater for each series of samples by plating sterile dilution water. Caution: Prepoured plates should non be contaminated past a surface colony or be below room temperature (water can well-up from agar). They should not be excessively dry, equally indicated by large wrinkles or glazed appearance. They should not accept water droplets on surface of agar or differences greater than 2 mm in agar depth, and they should not be stored at 0-four.4°C for longer than l calendar month. Reduced flow charge per unit through tubing indicates obstructions or fabric in organisation. To clear obstructions, remove valve from syringe, insert paw-held syringe with Luer fitting containing water, and apply pressure. Utilize booze rinse to remove residual material adhering to walls of arrangement. Dissolve accumulated residual with chromic acid. Rinse well afterwards cleaning.
- Counting grid
- Description. Use same counting grid for both 100 and 150 mm petri dishes. A mask is supplied for apply with 100 mm dishes. Counting grid is divided into 8 equal wedges; each wedge is divided by 4 arcs labeled fifty, 2, iii, and 4 from outside grid edge. Other lines within these arcs are added for ease of counting. A segment is the area between 2 arc lines within a wedge. Number of areas counted (e.1000., iii) means number of segments counted within a wedge. Screw plater deposits sample on agar plate in the same way each fourth dimension. The grid relates colonies on spiral plate to the volume in which they were contained. When colonies are counted with grid, sample volume becomes greater equally counting starts at outside edge of plate and proceeds toward center of plate.
- Calibration. The volume of sample represented by diverse parts of the counting grid is shown in operator's transmission that accompanies spiral plater. Grid expanse constants take been checked by the manufacturer and are accurate. To verify these values, gear up 11 bacterial concentrations in range of 106-103 cells/ml past making i:one dilutions of bacterial pause (use a nonspreader). Plate all Incubate both sets of plates for 48 ± 3 h at 35 ± 1°C. Summate concentrations for each dilution. Count spiral plates over grid surface, using counting rule of xx (described in H, below), and record number of colonies counted and filigree surface area over which they were counted. Each spiral colony count for a particular grid area, divided by aerobic count/ml for corresponding spirally plated bacterial concentrations, indicates volume deposited on that item filigree area. Employ the following formula:
To cheque total volume dispensed by spiral plater, weigh amount dispensed from stylus tip. Collect in tared 5 ml plastic beaker and weigh on analytical balance (± 0.2 mg).
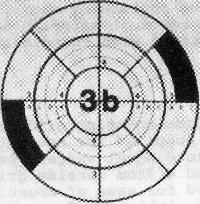
Figure one. 10 cm plate, area (3b)
- Examination and reporting of spiral plate counts.
Counting dominion of 20. Later on incubation, center screw plate over grid past adjusting holding artillery on viewer. Cull whatever wedge and begin counting colonies from outer edge of beginning segment toward eye until twenty colonies have been counted. Complete by counting remaining colonies in segment where 20th colony occurs. In this counting procedure, numbers such as 3b, 4c (Fig. l) refer to area segments from outer edge of wedge to designated arc line. Whatever count irregularities in sample composition are controlled by counting the same segments in the opposite wedge and recording results. Example of spirally inoculated plate (Fig. 50) demonstrates method for determining microbial count. Ii segments of each wedge were counted on reverse sides of plate with 31 and 30 colonies, respectively. The sample volume contained in the darkened segments is 0.0015 ml. To guess number of microorganisms, divide count by book contained in all segments counted. Meet instance nether Fig. l.
If 20 CFU are not within the 4 segments of the wedge, count CFU on entire plate. If the number of colonies exceeds 75 in second, third, or 4th segment, which also contains the 20th colony, the estimated number of microorganisms will mostly be low because of coincidence error associated with crowding of colonies. In this instance, count each circumferentially adjacent segment in all eight wedges, counting at least 50 colonies, e.g., if the outset two segments of a wedge incorporate 19 colonies and the third segment contains the 20th and 76th (or more), count colonies in all circumferentially adjacent outset and second segments in all 8 wedges. Calculate contained volume in counted segments of wedges and divide into number of colonies.
When fewer than xx colonies are counted on the total plate, report results as "less than 500 estimated SPLC per ml." If colony count exceeds 75 in first segment of wedge, report results every bit "greater than 500,000 estimated SPLC per ml." Do not count spiral plates with irregular distribution of colonies caused past dispensing errors. Report results of such plates as laboratory accident (LA). If spreader covers entire plate, discard plate. If spreader covers half of plate surface area, count just those colonies that are well distributed in spreader-costless areas.
Compute SPLC unless restricted by detection of inhibitory substances in sample, excessive spreader growth, or laboratory accidents. Round off counts equally described in I-D, to a higher place. Written report counts as SPLC or estimated SPLC per ml.
References
- American Public Health Association. 1984. Compendium of Methods for the Microbiological Examination of Foods, 2nd ed. APHA, Washington, DC
- American Public Wellness Clan. 1993. Standard Methods for the Test of Dairy Products, 16th ed. APHA, Washington, DC.
- Clan of Official Belittling Chemists. 1990. Official Methods of Analysis, 15th ed. AOAC, Arlington, VA.
- Donnelly, C.B., J.E. Gilchrist, J.T. Peeler, and J.E. Campbell. 1976. Spiral plate count method for the examination of raw and pasteurized milk. Appl. Environ. Microbiol. 32:21-27.
- Gilchrist, J.E., C.B. Donnelly, J.T. Peeler, and J.Eastward. Campbell. 1977. Collaborative written report comparing the spiral plate and aerobic plate count methods. J. Assoc. Off. Anal. Chem. 60:807-812.
- International Dairy Federation. 1987. Milk and Milk Products: Enumeration of Microorganisms—Colony Count at 3°C. Provisional IDF Standard 100A. IDF, Brussels, Belgium.
- Jarvis, B., V.H. Lach, and J.K. Wood. 1977. Evaluation of the spiral plate maker for the enumeration of microorganisms in foods. J. Appl. Bacteriol. 43:149-157.
- Niemela, S. 1983. Statistical evaluation of Results from Quantitative Microbiological Examinations. Study No. 1, 2d ed. Nordic Committee in Nutrient Analysis, Uppsala, Sweden.
- Tomasiewicz, D.K., D.Chiliad. Hotchkiss, Thou.W. Reinbold, R.B. Read, Jr., and P.A. Hartman. 1980. The most suitable number of colonies on plates for counting. J. Food Prot. 43:282-286.
- Zipkes, M.R., J.E. Gilchrist, and J.T. Peeler. 1981. Comparison of yeast and mold counts by spiral, cascade, and streak plate methods. J. Assoc. Off. Anal. Chem. 64:1465-1469.
Hypertext Source: Bacteriological Belittling Transmission, Edition 8, Revision A, 1998. Chapter three.
Source: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-3-aerobic-plate-count
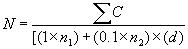
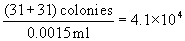
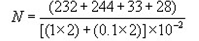
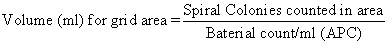

0 Response to "Standard Plate Count Microbiology Results Beef"
Post a Comment